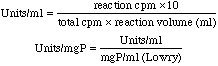Reverse Transcriptase, Recombinant HIV Assay
Unit definition: One unit incorporates 1 nanomole of tritiated dTMP into acid insoluble productsusing poly(A)•oligo(dT) 12-18 as the template-primer in 20 minutes at 37° C.
Reagents
Enzyme dilute as needed wtih 0.05M Tris/HCl, pH 8.3, 0.008M MgCl2 containing 0.1 mg/mL (1%) BSA Procedure Pipette into each tube as follows:
Incubate 20 minutes at 37° C. Stop reaction by adding 1 ml 10% cold perchloric acid. Filter through 0.2μ manifold filters used with Millipore vacuum manifold. Wash four times using 2mL 1% cold perchloric acid/wash. Transfer filter to scintillation vials. Add 2mL Cellosolve (or 2-methoxyethanol) to dissolve filter. Filters become opaque upon addition of Cellosolve. Make sure filters are dissolved before proceeding. Add 10mL scintillation cocktail and count. Calculations |

 Place Order
Place Order