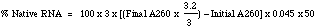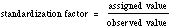Ribonucleic Acid Assay
Method: Determine suitability as substrate for RNase by using as substrate in RNase assay. Standardization of ribonuclease activity has been difficult due to varying rates at which reactions occur as well as to the significant differences in nucleotide patterns in RNA isolated from biological sources. Crook et al. (1960) have published an assay using a synthetic substrate, cytidine 2', 3'-phosphate. Zimmerman and Sandeen (1965) described a sensitive assay using polycytidylic acid.
The method of Kalnitsky et al. (1959) is used at Worthington. The rate of hydrolysis of yeast RNA at pH 5.0 is determined by measuring the amount of acid soluble oligonucleotide released under defined conditions. One unit causes an increase in absorbance of 1.0 at A260 at 37°C and pH 5.0 under the specified conditions. Reagents 0.10 M Sodium acetate buffer, pH 5.0: Dissolve 5.75 ml of glacial acetic acid in 900 ml reagent grade water. Adjust pH to 5.0 with 5 N NaOH and bring to a final volume of 1000 ml with reagent grade water. 25% Perchloric acid containing 0.75% Uranyl acetate: Dissolve 50 ml of 70% perchloric acid (HClO4) in 90 ml reagent grade water. Add 750 mg uranyl acetate (MW 424.15) and stir to dissolve. RNA Solution: Dissolve 100 mg Worthington Ribonucleic Acid (RNA) in 10 ml 0.1 M sodium acetate buffer, pH 5.0 Stir gently to dissolve to prevent denaturation. Record A260 at 1:50 dilution in 0.10M sodium acetate pH 5.0 to determine mg RNA/ml. mg RNA/ml = A260 x dilution x 0.04 RNAse Immediately prior to assay, dilute further to 2, 4, and 6 micrograms per milliliter in 0.10 M sodium acetate, pH 5.0. Procedure Determine % Native RNA: Dissolve 10 mg RNA in 10 ml 0.015% NaCl. Dilute 1:50 in NaCl and pipette 3 ml into cuvette and read at 260 nm and 320 nm. Add 0.2 ml 5N NaOH (Fresh, stored in plastic). Read at 260 and 320 nm. Stand one hour at 37°C. Read at 260 and 320 nm. Determine % Native: (Contact supervisor if less than 60%.).A320 is a check for extraneous background. Calculation of % Native RNA:
RNase Blank Assay: (Can be set up with RNase assay) Set up a blank for sample RNA and a blank for test RNA. Pipet one ml of 0.10M sodium acetate buffer pH 5.0 into centrifuge tubes. Equilibrate to 37°C. Add one ml of RNA Solution (2.5 mg/ml). Let set one hour at 37°C. Then add one ml of uranyl acetate-perchloric acid solution. Transfer to an ice bath and cool for 5 minutes. Clarify by centrifugation and dilute 0.1ml of clear supernatatnt to 3.0ml with reagent grade water. Read A260 vs blank. Blank should not increase more than 50% in one hour at 37°C. RNase Assay: Set up one set of tubes for Sample and one for Control. Include a Blank with the Sample set of tubes, and a Blank with the Control set of tubes. Pipette one ml of respective enzyme dilution into centrifuge tubes; pipette one ml of 0.10M sodium acetate buffer pH 5.0 into Blank tubes. Incubate all tubes at 37°C for 58 minutes. At timed intervals, add one ml of RNA Solution (2.5 mg/ml) to all tubes (Samples, Controls, and Blanks). Incubate each tube exactly 4 minutes and stop reaction by the addition of one ml of uranyl acetate-perchloric acid solution. Transfer to an ice bath and cool for 5 minutes. Clarify by centrifugation and dilute 0.1 ml of clear supernatant to 3.0 ml with reagent grade water. Read A260 versus blank. Calculation Note: Note: Because the RNA used as a substrate is derived from a natural source, lot to lot variation occurs. To overcome this, a ribonuclease standard is run and values are adjusted to the standard. Example: If the value assigned to the standard is 2500 u/mg and the value obtained for the standard is 2000 u/mg, then a factor of 1.25 is to be applied to the assay values. |

 Place Order
Place Order